-
Research
-
Publications
-
All publications
-
Benner, SA
-
Biondi, E
-
Bradley, K
-
Chen, C
-
Hoshika, S
-
Karalkar, N
-
Kim, HJ
-
Kim, MJ
-
Laos, R
-
Leal, NA
-
Li, Y
-
Shaw, RW
-
Spacek, J
-
Yang, ZY
-
People
-
Benner, Steven
-
Biondi, Elisa
-
Bradley, Kevin
-
Chen, Cen
-
Darling, April
-
Hoshika, Shuichi
-
Karalkar, Nilesh
-
Kim, Hyo-Joong
-
Kim, Myong-Jung
-
Laos, Roberto
-
Leal, Nicole
-
Li, Yubing
-
Shaw, Ryan
-
Spacek, Jan
-
Yang, Zunyi
-
News and Events
-
Press Coverage
-
Our Foundation
|
Synthetic Biology

Re-inventing Proteins
The availability of extra letters in the genetic alphabet projects allows DNA and RNA to encode proteins with additional amino acids. This allows us to re-invent proteins. A team at the Foundation is now doing this for proteins that contain post-translational modifications, such as acetylation and phosphorylation. This work requires the engineering of nucleic acids, but also enzymes that charge nucleic acids with amino acids, and molecular assemblies that convert information in DNA and RNA into proteins. This work is now supported by the National Cancer Institute.
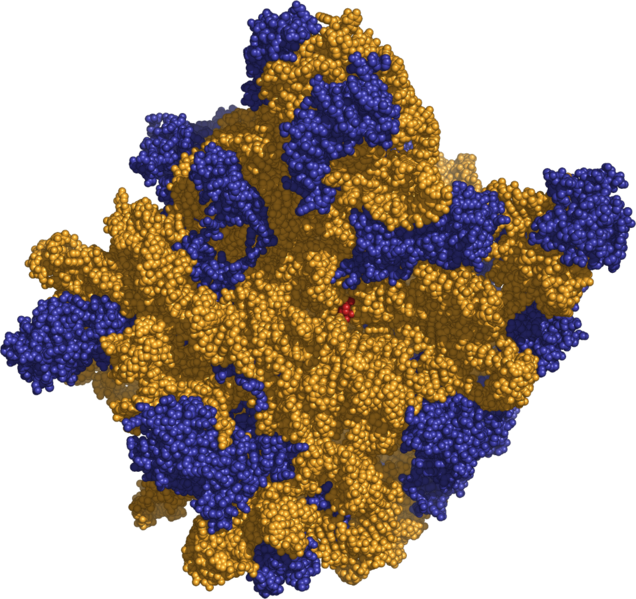
The ribosome, with its RNA component (orange), can synthesize proteins with extra amino acids
using extra codons built from the extra nucleotides in the AEGIS DNA alphabet.

Evolving receptors, ligands, and catalysts
In addition to being a platform to understand the science of molecules that hold biological information, its transfer, and its evolution, AEGIS is itself able to evolve under selection pressure to perform function. A decade of work in the laboratory has developed polymerases that copy AEGIS DNA, that make AEGIS RNA from AEGIS DNA, and make AEGIS DNA from AEGIS RNA. A team at FfAME developed technology to sequence AEGIS DNA. This has allowed Foundation scientists to create AEGISzymes that bind selectively to cancer cells, inactivate toxins, and catalyze reactions. These have broad use in research, diagnostics, and therapeutics.
Researchers at FfAME are now supported by the National Institutes of Health to develop this technology platform. Another research program is being supported by the National Institute of General Medical Sciences of the NIH to apply this technology in cancer research. Further, a collaboration with the University of Washington is developing next generation tools to sequence AEGIS DNA using nanopores. This collaboration is presently funded by the National Human Genome Research Institute.

Crystals of an AEGISbody evolved from a 6 letter
alphabet to bind to an anthrax target.

Biotechnology, DNA and RNA synthesis and sequencing
The redesign of DNA, RNA, and proteins helps us understand the molecular science that stands behind biology, evolution, and natural history. It also is the foundation for biotechnology that supports research in molecular biology, as well as medical diagnostics and therapeutics.
For example, in 1984, the Benner laboratory was the first to synthesize a DNA molecule to encode an enzyme. This work introduced tactics now standard in synthetic biology, including optimized codons, restriction sites to facilitate downstream engineering, and watermarks to allow the provenance of a synthetic DNA genetic construct to be determined.
Our group also introduced the "aminoxy" reversible terminating group to support sequencing and enzymatic synthesis of DNA. This has been licensed for enzyme-based DNA synthesis at DNA Script. The same aminoxy moiety is licensed for DNA sequencing, including personal sequencing, and single molecule sequencing. FfAME researchers are using this group to develop tools for the enzymic synthesis of RNA, in work now supported by the National Human Genome Research Institute.
To make ultra-large constructs of DNA, our group invented architectures that exploit AEGIS and various enzyme systems that manipulate them. This ongoing research also seeks to prepare DNA having extremely high purity and fidelity, to allow one pot autonomous self-assembly of complete plasmids and genes. These projects are now supported by the Department of Energy through its bioeconomy synthetic biology program.
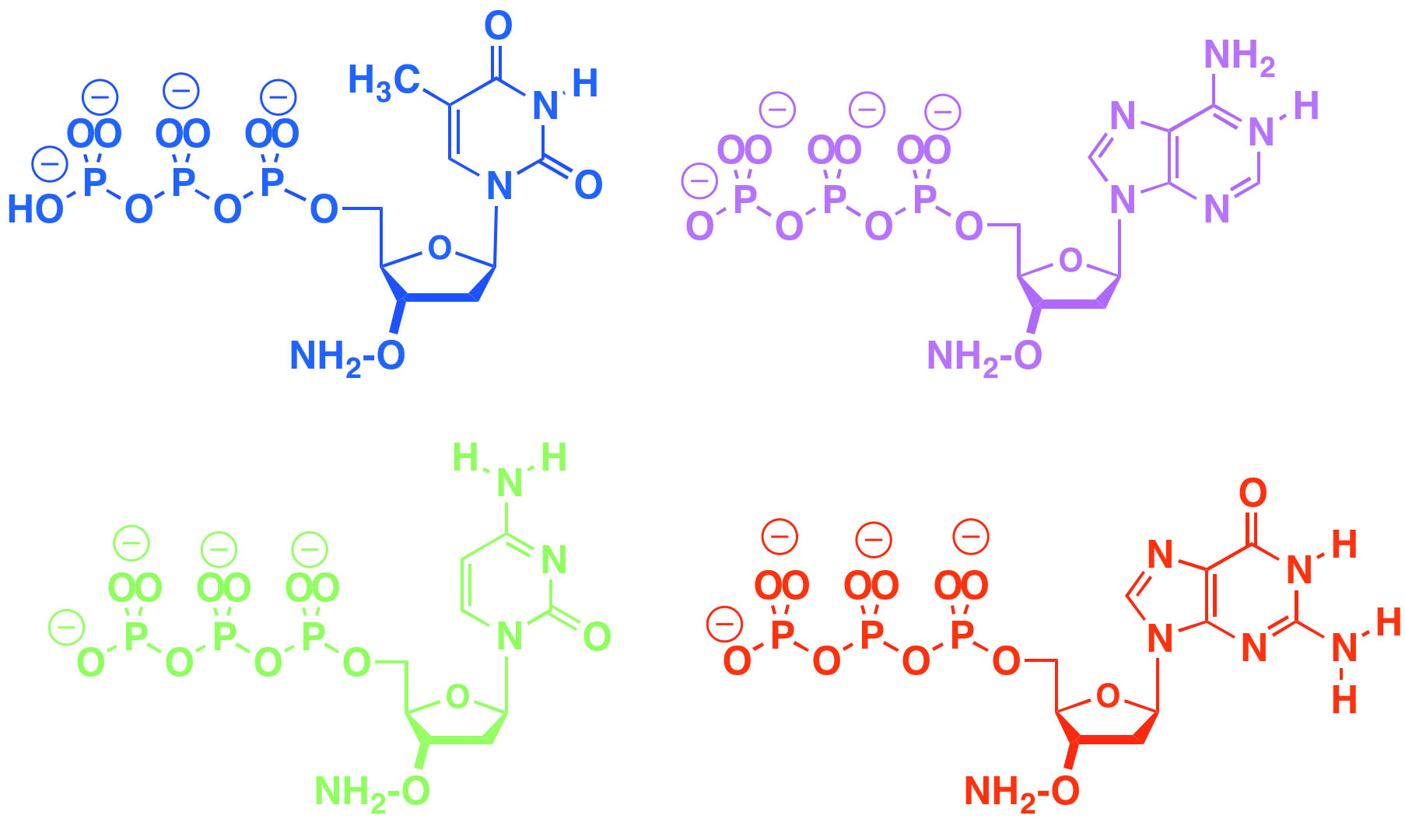
Aminoxy triphosphates support both next generation DNA sequencing and enzyme-based DNA and RNA synthesis by allowing an enzyme to add them to a growing DNA primer, stop, give time for analysis and deblocking, and then repeat.

Re-inventing pathways
Another theme in synthetic biology matches enzymes from different biosources to create new pathways. Our group contributed to the development of metabolic engineering, for example, by developing engineered metabolic pathways that exploit thermodynamic features favored in natural pathways. The next horizon in synthetic biology moves these pathways into living cells. An ongoing project, supported by the National Science Foundation, seeks to install pathways to allow living cells to exploit AEGIS DNA. This project is now supported by the National Science Foundation.
A still higher goal in cell design avoids the use of a natural cell entirely. To this end, artificial cells are also being developed based on fluorinated oils and surfactants invented at the Foundation
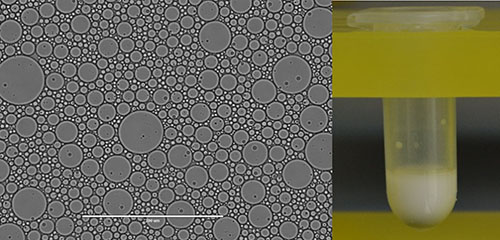
An image of artificial cells made with fluorinated oils and surfactants
|
|
|



